Data Analysis(2)
The data is available here
The source code is available here
Instruction
- The data set is about the abalone.
-
Number of examples: 4177
- Predicting the age of abalone from physical measurements.
- The age of abalone is determined by cutting the shell through the cone, staining it, and counting the number of rings through a microscope – a boring and time-consuming task.
- Other measurements, which are easier to obtain, are used to predict the age.
- Further information, such as weather patterns and location (hence food availability) may be required to solve the problem.
Attributes
Given is the attribute name, attribute type, the measurement unit and a
brief description. The number of rings is the value to predict: either
as a continuous value or as a classification problem.
| Name | Data Type | Meas. | Description |
|---|---|---|---|
| Sex | nominal | None | M, F, and I (infant) |
| Length | continuous | mm | Longest shell measurement |
| Diameter | continuous | mm | perpendicular to length |
| Height | continuous | mm | with meat in shell |
| Whole weight | continuous | grams | whole abalone |
| Shucked weight | continuous | grams | weight of meat |
| Viscera weight | continuous | grams | gut weight (after bleeding) |
| Shell weight | continuous | grams | after being dried |
| Rings | integer | +1.5 | gives the age in years |
Statistics for numeric domains:
| Attr | Length | Diam | Height | Whole | Shucked | Viscera | Shell | Rings |
|---|---|---|---|---|---|---|---|---|
| Min | 0.075 | 0.055 | 0.000 | 0.002 | 0.001 | 0.001 | 0.002 | 1 |
| Max | 0.815 | 0.650 | 1.130 | 2.826 | 1.488 | 0.760 | 1.005 | 29 |
| Mean | 0.524 | 0.408 | 0.140 | 0.829 | 0.359 | 0.181 | 0.239 | 9.934 |
| SD | 0.120 | 0.099 | 0.042 | 0.490 | 0.222 | 0.110 | 0.139 | 3.224 |
| Correl | 0.557 | 0.575 | 0.557 | 0.540 | 0.421 | 0.504 | 0.628 | 1.0 |
Class Distribution:
| No. | Class | Examples | Class | Examples |
|---|---|---|---|---|
| 1 | 1 | 2 | 1 | |
| 3 | 15 | 4 | 57 | |
| 5 | 115 | 6 | 259 | |
| 7 | 391 | 8 | 568 | |
| 9 | 689 | 10 | 634 | |
| 11 | 487 | 12 | 267 | |
| 13 | 203 | 14 | 126 | |
| 15 | 103 | 16 | 67 | |
| 17 | 58 | 18 | 42 | |
| 19 | 32 | 20 | 26 | |
| 21 | 14 | 22 | 6 | |
| 23 | 9 | 24 | 2 | |
| 25 | 1 | 26 | 1 | |
| 27 | 2 | 29 | 1 |
Total 4177
Preprocessing
- For the
Sexcolumn, i prefer on-hot vectorization to setting three numbers forM,F,I - For the
Ringscolumn, i didn’t add 1.5 to it to get age but using the raw data instead
import pandas as pd
import numpy as np
data = pd.read_csv("./abalone_data.txt", names=["Sex", "Length", "Diamter", "Height",
"WholeWeight", "ShuckedWeight", "VisceraWeight", "SellWeight", "Rings"])
# data.replace(to_replace="M", value="1", inplace=True)
# data.replace(to_replace="F", value="0", inplace=True)
# data.replace(to_replace="I", value="0.5", inplace=True)
Sex = data.pop("Sex")
I = Sex.copy()
I.replace(to_replace="I", value=1, inplace=True)
I.replace(to_replace="M", value=0, inplace=True)
I.replace(to_replace="F", value=0, inplace=True)
M = Sex.copy()
M.replace(to_replace="I", value=0, inplace=True)
M.replace(to_replace="M", value=1, inplace=True)
M.replace(to_replace="F", value=0, inplace=True)
F = Sex.copy()
F.replace(to_replace="I", value=0, inplace=True)
F.replace(to_replace="M", value=0, inplace=True)
F.replace(to_replace="F", value=1, inplace=True)
data.insert(0, "I", I)
data.insert(0, "F", F)
data.insert(0, "M", M)
print(data.columns)
print(data.corr())
Index(['M', 'F', 'I', 'Length', 'Diamter', 'Height', 'WholeWeight',
'ShuckedWeight', 'VisceraWeight', 'SellWeight', 'Rings'],
dtype='object')
M F I Length Diamter Height \
M 1.000000 -0.512528 -0.522541 0.236543 0.240376 0.215459
F -0.512528 1.000000 -0.464298 0.309666 0.318626 0.298421
I -0.522541 -0.464298 1.000000 -0.551465 -0.564315 -0.518552
Length 0.236543 0.309666 -0.551465 1.000000 0.986812 0.827554
Diamter 0.240376 0.318626 -0.564315 0.986812 1.000000 0.833684
Height 0.215459 0.298421 -0.518552 0.827554 0.833684 1.000000
WholeWeight 0.252038 0.299741 -0.557592 0.925261 0.925452 0.819221
ShuckedWeight 0.251793 0.263991 -0.521842 0.897914 0.893162 0.774972
VisceraWeight 0.242194 0.308444 -0.556081 0.903018 0.899724 0.798319
SellWeight 0.235391 0.306319 -0.546953 0.897706 0.905330 0.817338
Rings 0.181831 0.250279 -0.436063 0.556720 0.574660 0.557467
WholeWeight ShuckedWeight VisceraWeight SellWeight Rings
M 0.252038 0.251793 0.242194 0.235391 0.181831
F 0.299741 0.263991 0.308444 0.306319 0.250279
I -0.557592 -0.521842 -0.556081 -0.546953 -0.436063
Length 0.925261 0.897914 0.903018 0.897706 0.556720
Diamter 0.925452 0.893162 0.899724 0.905330 0.574660
Height 0.819221 0.774972 0.798319 0.817338 0.557467
WholeWeight 1.000000 0.969405 0.966375 0.955355 0.540390
ShuckedWeight 0.969405 1.000000 0.931961 0.882617 0.420884
VisceraWeight 0.966375 0.931961 1.000000 0.907656 0.503819
SellWeight 0.955355 0.882617 0.907656 1.000000 0.627574
Rings 0.540390 0.420884 0.503819 0.627574 1.000000
Visualization
- Some preprocessing won’t be done before the visualization as shown below.
# count the numbers of every class, which is included in the instruction.
# print(data.dtypes)
counts = []
for i in range(1, 30):
count = data.loc[lambda df: df["Rings"] == float(i)]
counts.append(count.shape[0])
print(counts)
[1, 1, 15, 57, 115, 259, 391, 568, 689, 634, 487, 267, 203, 126, 103, 67, 58, 42, 32, 26, 14, 6, 9, 2, 1, 1, 2, 0, 1]
%matplotlib inline
import matplotlib.pyplot as plt
plt.bar(range(1, 30), counts)
plt.show()
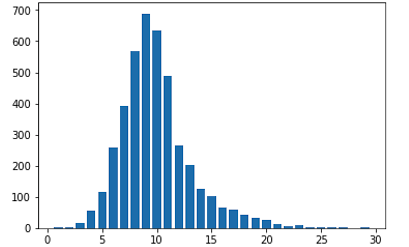
- As you see the class distribution looks like the Gauss distribution, which we need to seperate the class like that
def ManualMap(x):
array = [1, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 16, 29]
for i in range(len(array)):
if x <= array[0]:
return 0
if array[i] <= x <= array[i + 1]:
return i + 1
if x >= array[-1]:
return len(array)
def LogMap(x):
array = np.around(np.geomspace(1, 29, 13))
for i in range(len(array)):
if x <= array[0]:
return 0
if array[i] <= x <= array[i + 1]:
return i + 1
if x >= array[-1]:
return len(array)
d1 = pd.DataFrame(data["Rings"].map(ManualMap)) #[i for i in map(fun, data["Rings"])
# print(d1.shape, d1.columns)
counts = []
for i in range(1, 30):
count = d1.loc[lambda df: df["Rings"] == i].shape[0]
if count != 0:
counts.append(count)
plt.subplot("121")
plt.bar(range(len(counts)), counts)
plt.xticks(range(len(counts)))
plt.xlabel("Seperate By ManualMap")
data["Rings"] = data["Rings"].map(LogMap) #[i for i in map(fun, data["Rings"])
counts = []
for i in range(1, 30):
count = data.loc[lambda df: df["Rings"] == i].shape[0]
if count != 0:
counts.append(count)
plt.subplot("122")
plt.bar(range(len(counts)), counts)
plt.xticks(range(len(counts)))
plt.xlabel("Seperate By LogMap")
plt.tight_layout()
plt.show()
#data["Rings"] = d1
data = data.values.astype(np.float)
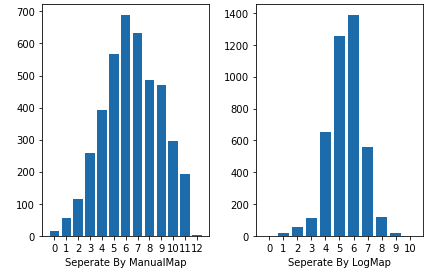
- Here you can see that with
ManualMapseperating the classes into 13 classes, and the result will be disperse. - While divide the class into 13 classes with
LogMap, it looks more likely the Gauss distribution.
Model builting
from sklearn.svm import SVC
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.ensemble import GradientBoostingClassifier
mms = MinMaxScaler()
mms.fit_transform(data[:, :-1])
X_train, X_test, y_train, y_test = train_test_split(data[:, :-1], data[:, -1], test_size=0.3)
def getScore(clf, X_test, y_test):
yPre = np.round(clf.predict(X_test))
# print(np.hstack((yPre, y_test.reshape(X_test.shape[0], 1))))
acc = (yPre == y_test)
print(f"{np.mean(acc) * 100:*^20}")
clf = SVC(gamma=10)
#clf = GradientBoostingClassifier(n_estimators=50, max_depth=2, learning_rate=0.5)
clf.fit(X_train, y_train)
getScore(clf, X_test, y_test)
getScore(clf, X_train, y_train)
print(cross_val_score(clf, X_test, y_test))
*54.066985645933016*
*54.567225453301404*
[0.5452381 0.51913876 0.56971154]
- It turns out that seperate into 13 classes is still to many for the classifiers to work with.You can try the 3-classes method to retrain the model, and the accuracy will be better than 80%.
- According to the offical suggestion, KNN and SVM working on the raw data, which has 20+ classes, get poor result.
- BTW: If you want to get better result try manualy seperate with array[1, 3 ,5, 8, 11, 29]
Summary
- The data sets beat me at the first place.For a long time, i have been working on Iris data set, which many classifier can do quite a good job.
- It reminded me of the importance of the accuracy of the training set and
cross_validationresult